
Extract summary from ulsif object, including two-sample significance test for homogeneity of the numerator and denominator samples
Source: R/summary.R
summary.ulsif.RdExtract summary from ulsif object, including two-sample significance
test for homogeneity of the numerator and denominator samples
Usage
# S3 method for class 'ulsif'
summary(
object,
test = FALSE,
n_perm = 100,
parallel = FALSE,
cluster = NULL,
...
)Arguments
- object
Object of class
ulsif- test
logical indicating whether to statistically test for homogeneity of the numerator and denominator samples.
- n_perm
Scalar indicating number of permutation samples
- parallel
logicalindicating to run the permutation test in parallel- cluster
NULLor a cluster object created bymakeCluster. IfNULLandparallel = TRUE, it uses the number of available cores minus 1.- ...
further arguments passed to or from other methods.
Examples
set.seed(123)
# Fit model
dr <- ulsif(numerator_small, denominator_small)
# Inspect model object
dr
#>
#> Call:
#> ulsif(df_numerator = numerator_small, df_denominator = denominator_small)
#>
#> Kernel Information:
#> Kernel type: Gaussian with L2 norm distances
#> Number of kernels: 150
#> sigma: num [1:10] 0.711 1.08 1.333 1.538 1.742 ...
#>
#> Regularization parameter (lambda): num [1:20] 1000 483.3 233.6 112.9 54.6 ...
#>
#> Optimal sigma (loocv): 1.538158
#> Optimal lambda (loocv): 2.976351
#> Optimal kernel weights (loocv): num [1:151] 0.0666 0.0289 0.0423 0.0442 0.0454 ...
#>
# Obtain summary of model object
summary(dr)
#>
#> Call:
#> ulsif(df_numerator = numerator_small, df_denominator = denominator_small)
#>
#> Kernel Information:
#> Kernel type: Gaussian with L2 norm distances
#> Number of kernels: 150
#>
#> Optimal sigma: 1.538158
#> Optimal lambda: 2.976351
#> Optimal kernel weights: num [1:151] 0.0666 0.0289 0.0423 0.0442 0.0454 ...
#>
#> Pearson divergence between P(nu) and P(de): 0.3868
#> For a two-sample homogeneity test, use 'summary(x, test = TRUE)'.
#>
# Plot model object
plot(dr)
#> Warning: Negative estimated density ratios for 2 observation(s) converted to 0.01 before applying logarithmic transformation
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
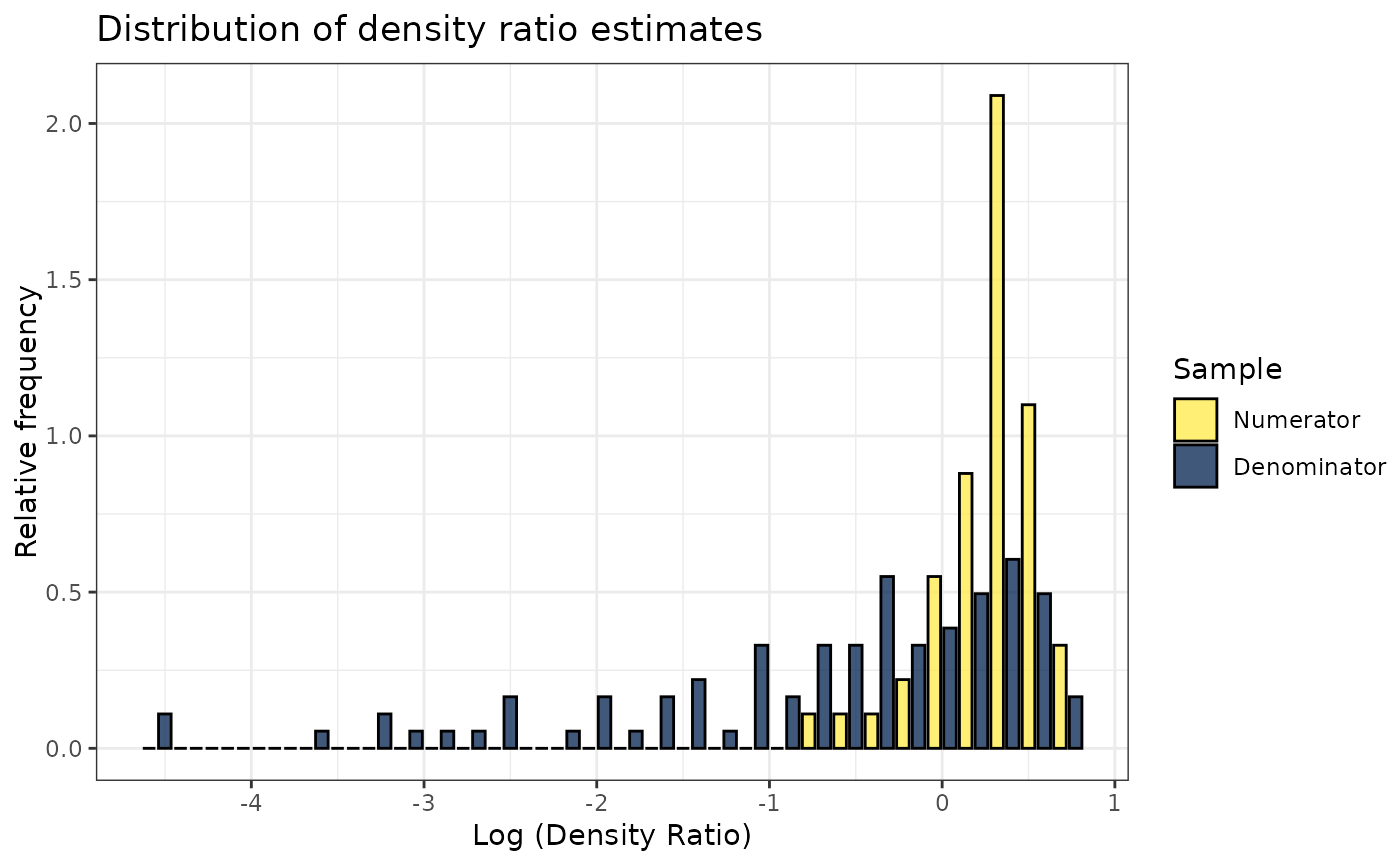 # Plot density ratio for each variable individually
plot_univariate(dr)
#> Warning: Negative estimated density ratios for 2 observation(s) converted to 0.01 before applying logarithmic transformation
#> [[1]]
# Plot density ratio for each variable individually
plot_univariate(dr)
#> Warning: Negative estimated density ratios for 2 observation(s) converted to 0.01 before applying logarithmic transformation
#> [[1]]
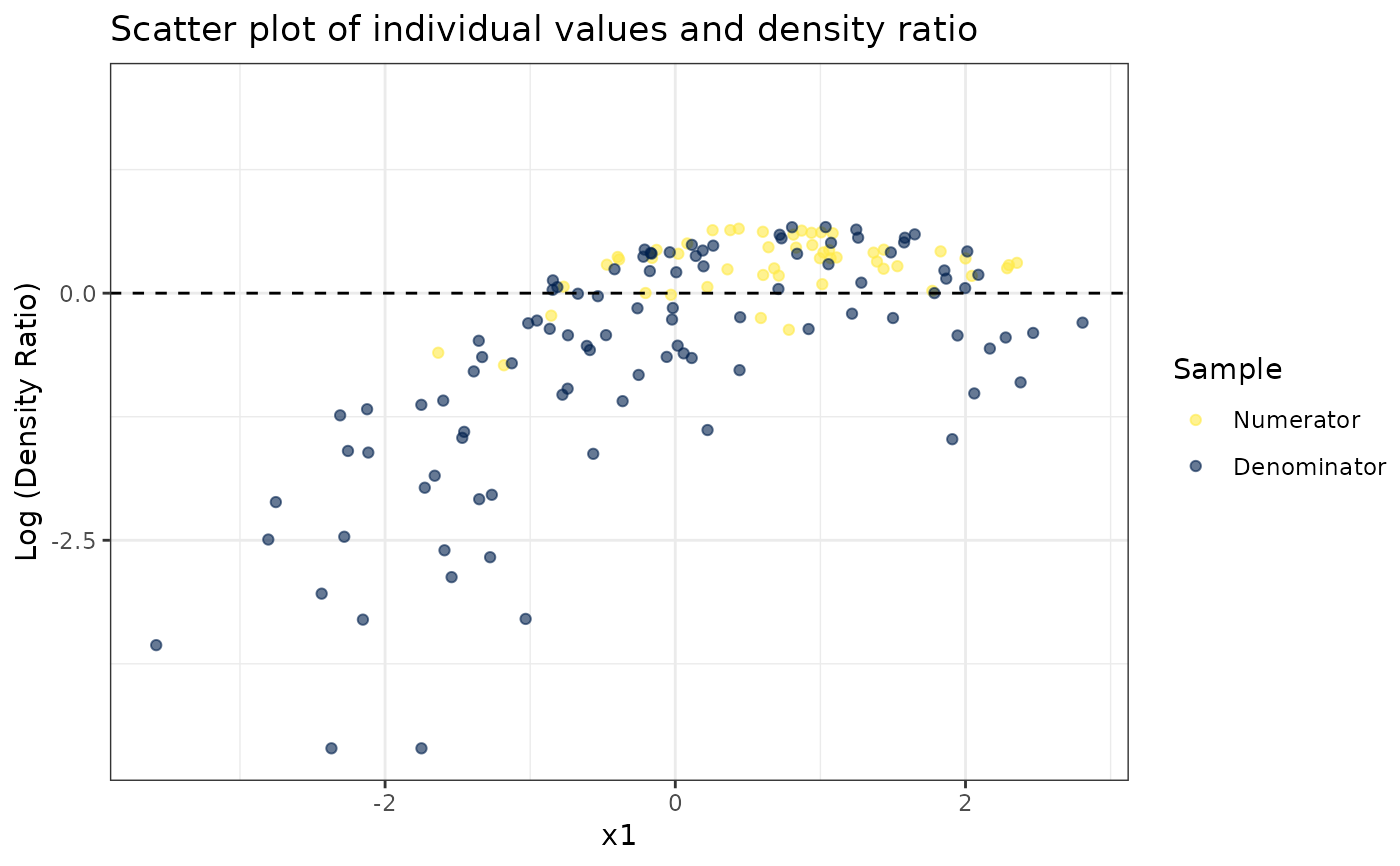 #>
#> [[2]]
#>
#> [[2]]
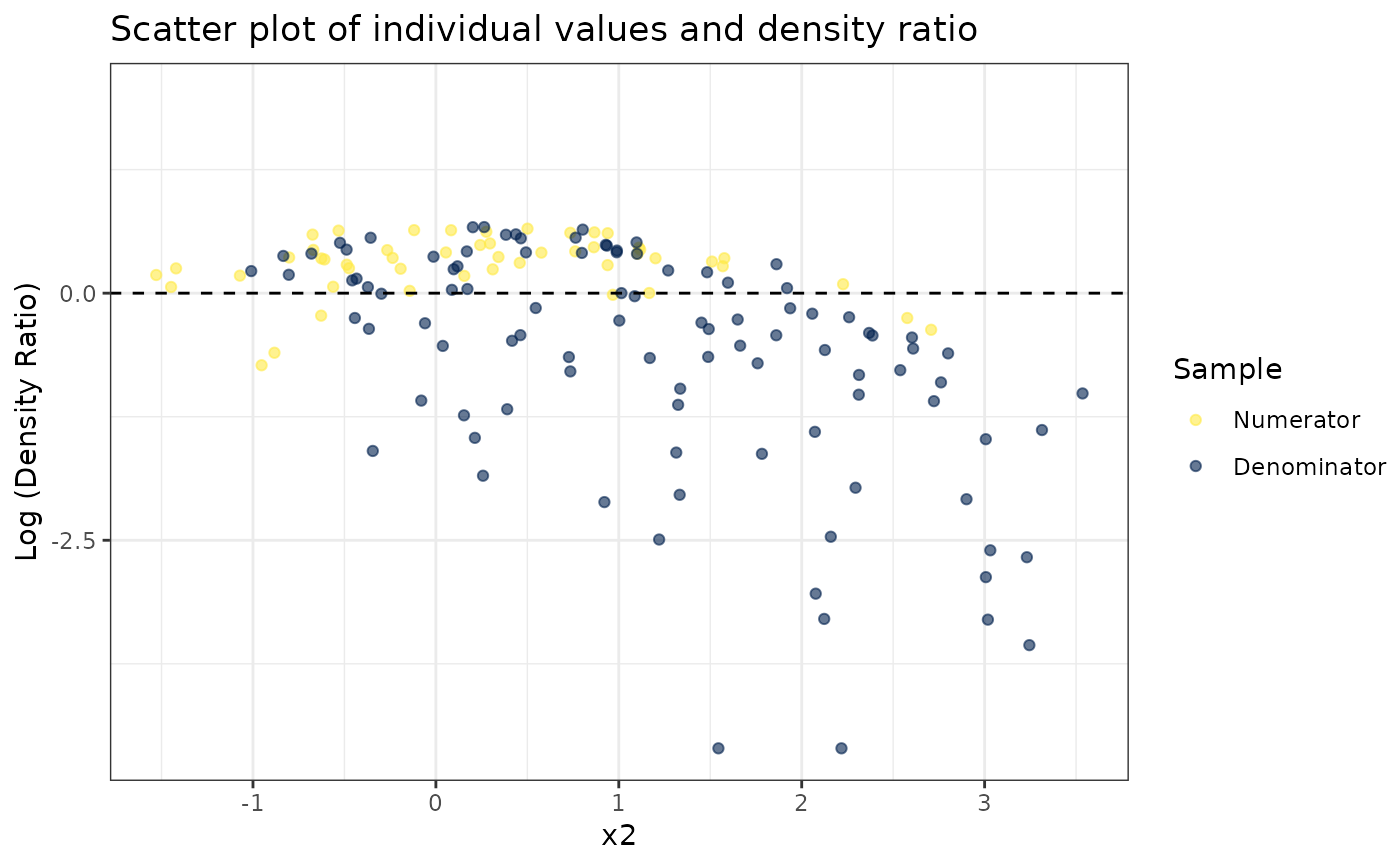 #>
#> [[3]]
#>
#> [[3]]
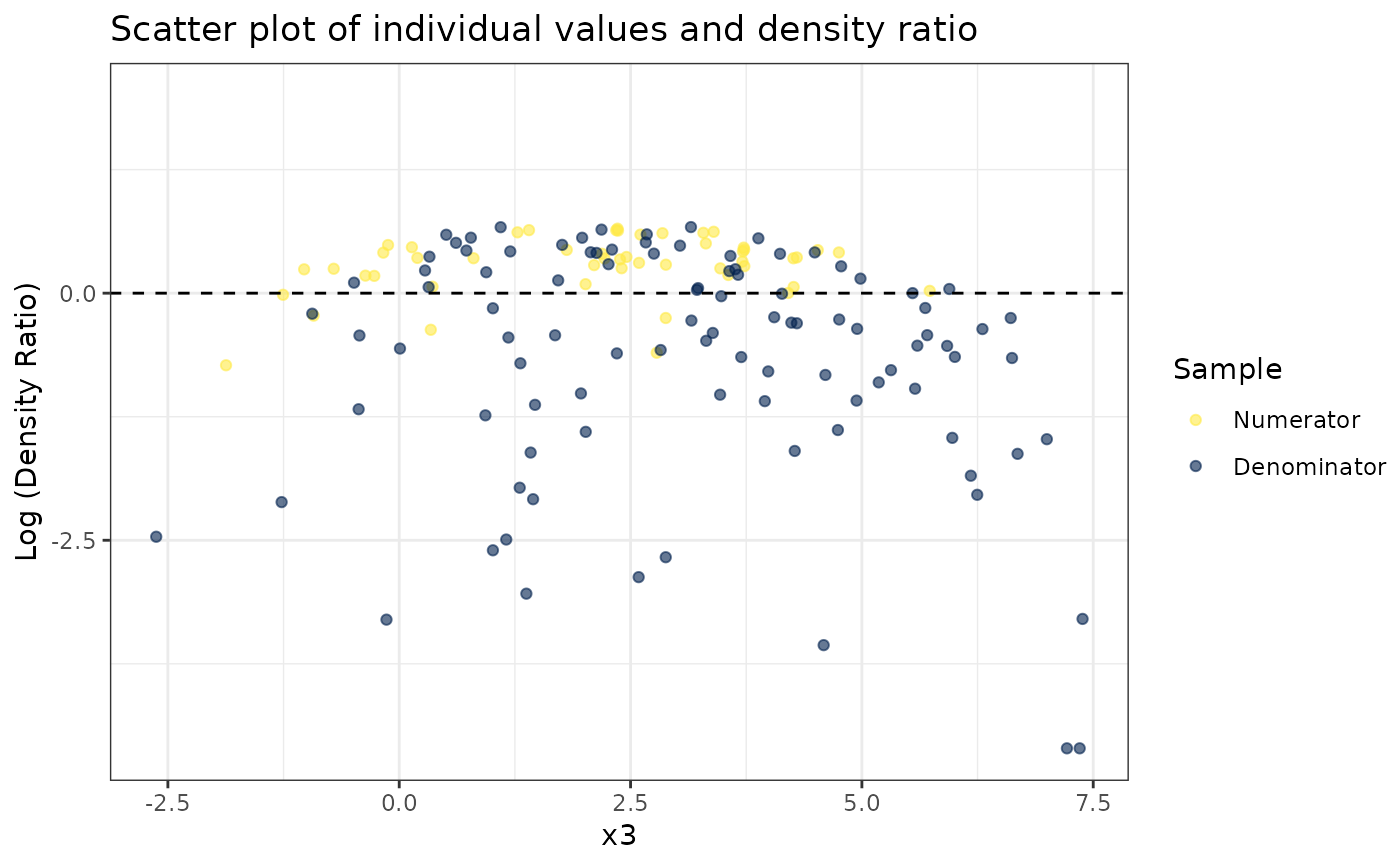 #>
# Plot density ratio for each pair of variables
plot_bivariate(dr)
#> Warning: Negative estimated density ratios for 2 observation(s) converted to 0.01 before applying logarithmic transformation
#> [[1]]
#>
# Plot density ratio for each pair of variables
plot_bivariate(dr)
#> Warning: Negative estimated density ratios for 2 observation(s) converted to 0.01 before applying logarithmic transformation
#> [[1]]
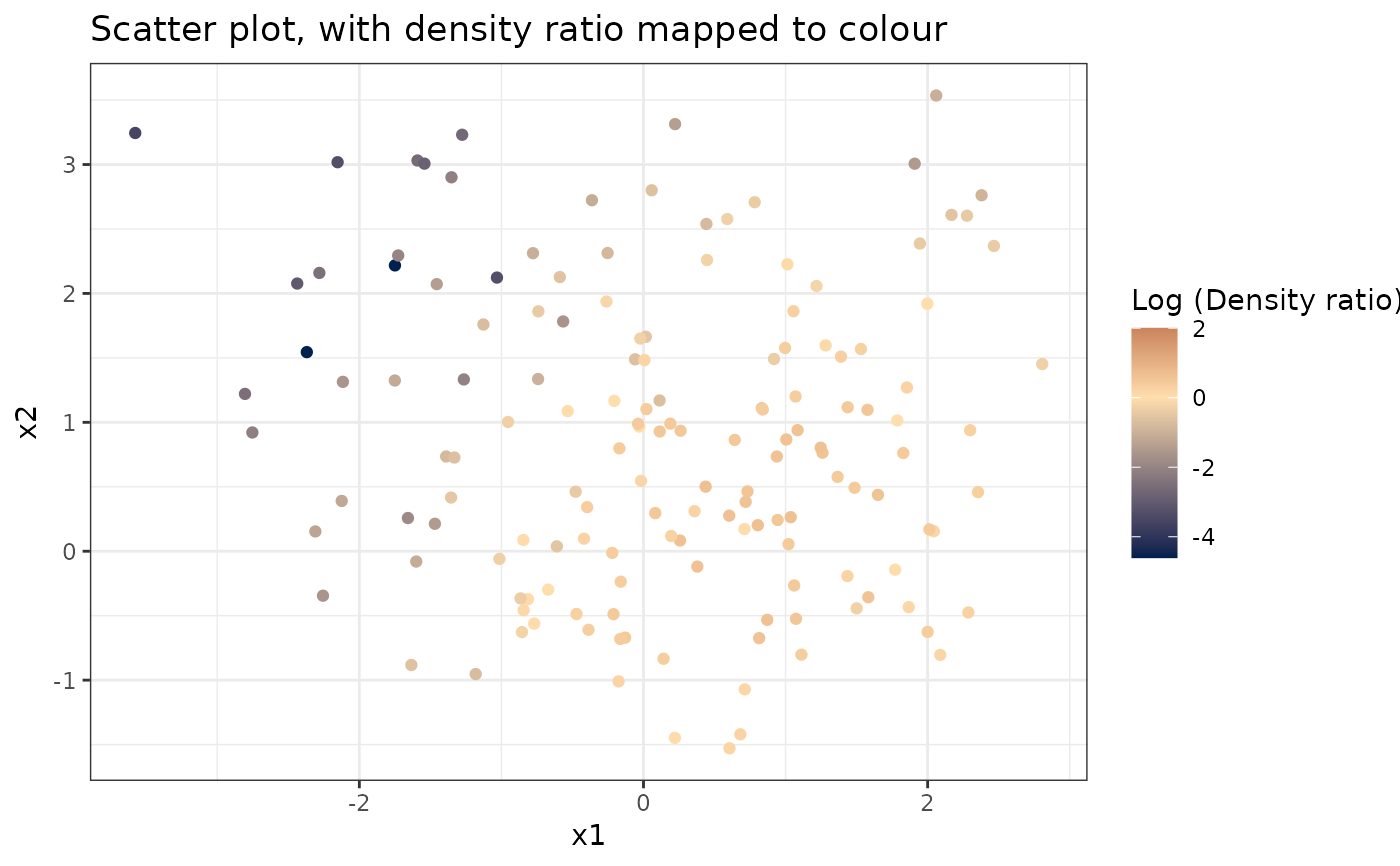 #>
#> [[2]]
#>
#> [[2]]
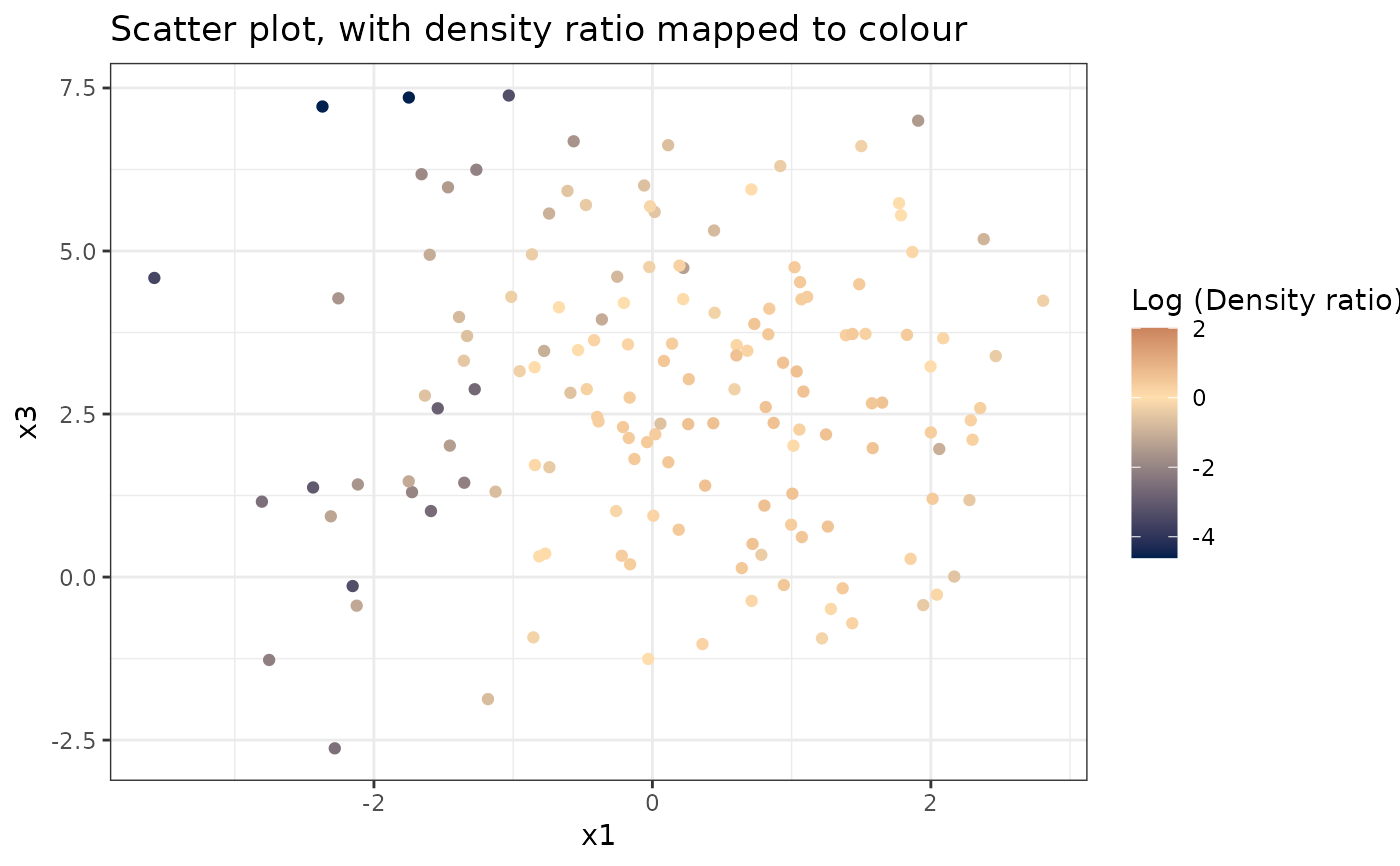 #>
#> [[3]]
#>
#> [[3]]
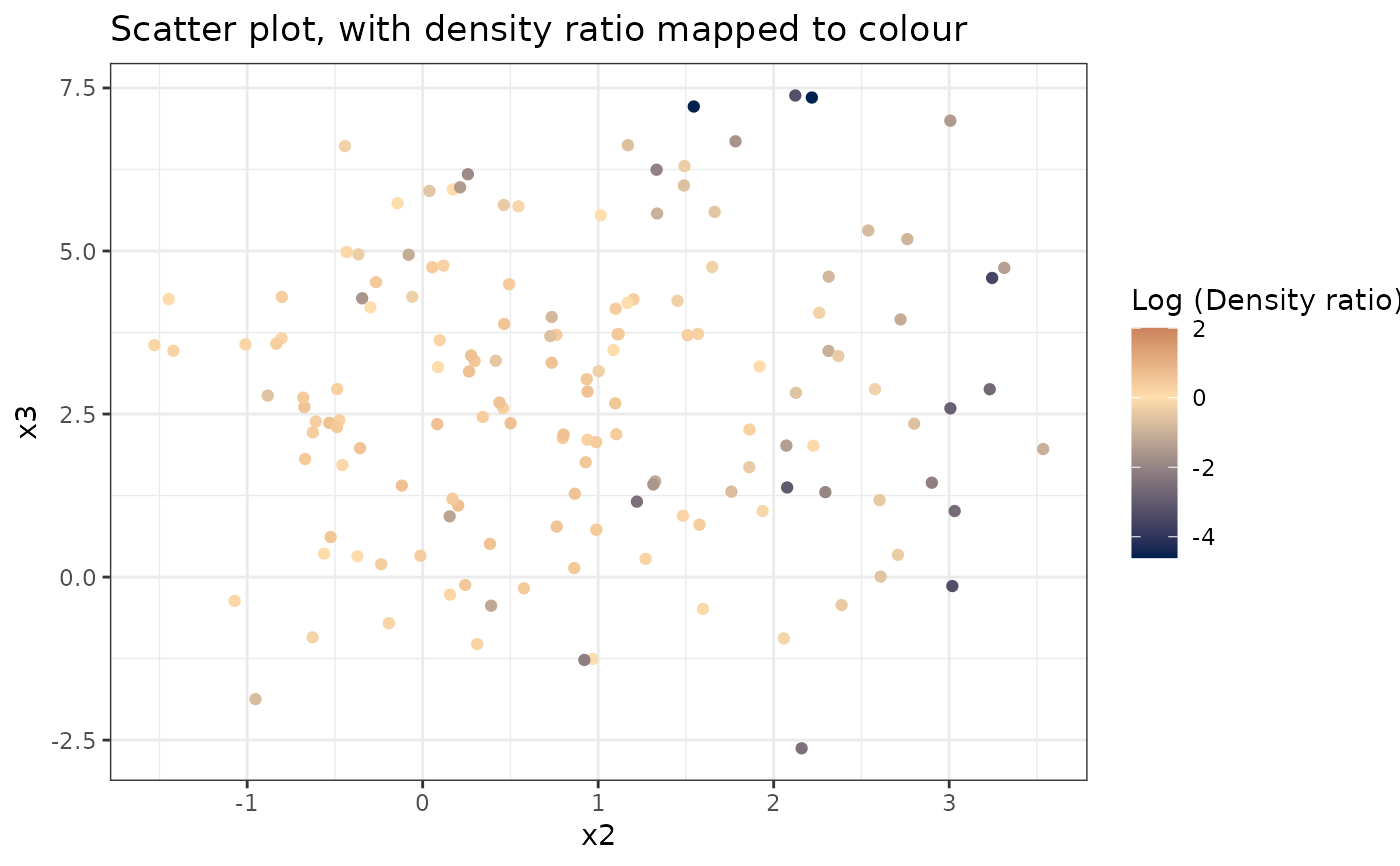 #>
# Predict density ratio and inspect first 6 predictions
head(predict(dr))
#> , , 1
#>
#> [,1]
#> [1,] 0.9838195
#> [2,] 1.2872509
#> [3,] 1.5069634
#> [4,] 1.2804095
#> [5,] 1.0953012
#> [6,] 1.5262485
#>
# Fit model with custom parameters
ulsif(numerator_small, denominator_small, sigma = 2, lambda = 2)
#>
#> Call:
#> ulsif(df_numerator = numerator_small, df_denominator = denominator_small, sigma = 2, lambda = 2)
#>
#> Kernel Information:
#> Kernel type: Gaussian with L2 norm distances
#> Number of kernels: 150
#> sigma: num 2
#>
#> Regularization parameter (lambda): num 2
#>
#> Optimal sigma (loocv): 2
#> Optimal lambda (loocv): 2
#> Optimal kernel weights (loocv): num [1:151] 0.0378 0.0348 0.0554 0.053 0.0619 ...
#>
#>
# Predict density ratio and inspect first 6 predictions
head(predict(dr))
#> , , 1
#>
#> [,1]
#> [1,] 0.9838195
#> [2,] 1.2872509
#> [3,] 1.5069634
#> [4,] 1.2804095
#> [5,] 1.0953012
#> [6,] 1.5262485
#>
# Fit model with custom parameters
ulsif(numerator_small, denominator_small, sigma = 2, lambda = 2)
#>
#> Call:
#> ulsif(df_numerator = numerator_small, df_denominator = denominator_small, sigma = 2, lambda = 2)
#>
#> Kernel Information:
#> Kernel type: Gaussian with L2 norm distances
#> Number of kernels: 150
#> sigma: num 2
#>
#> Regularization parameter (lambda): num 2
#>
#> Optimal sigma (loocv): 2
#> Optimal lambda (loocv): 2
#> Optimal kernel weights (loocv): num [1:151] 0.0378 0.0348 0.0554 0.053 0.0619 ...
#>